Simulating the Distance Distribution between Spin-Labels Attached to Proteins
Von einem Mystery-Man-Autor
Last updated 20 Juni 2024
Molecules, Free Full-Text

Ultrafast Bioorthogonal Spin-Labeling and Distance Measurements in Mammalian Cells Using Small, Genetically Encoded Tetrazine Amino Acids
Frontiers Exploring intrinsically disordered proteins using site-directed spin labeling electron paramagnetic resonance spectroscopy

EPR Distance Measurements on Long Non‐coding RNAs Empowered by Genetic Alphabet Expansion Transcription - Domnick - 2020 - Angewandte Chemie International Edition - Wiley Online Library

Cross-validation of distance measurements in proteins by PELDOR/DEER and single-molecule FRET

Molecules, Free Full-Text

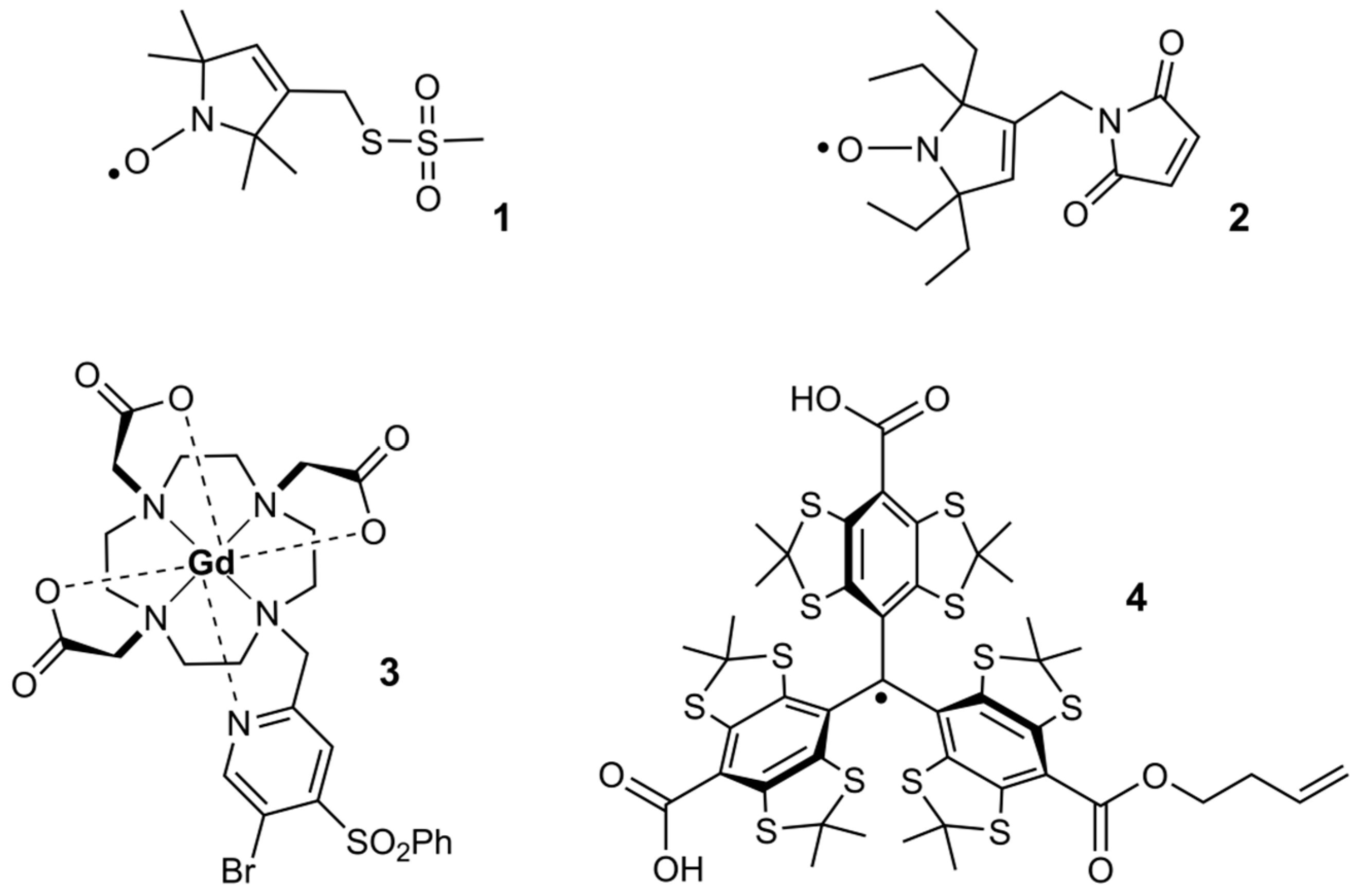
Spin-labeled nucleosides. (a) Rigid spin labels Ç and Ç m (b)

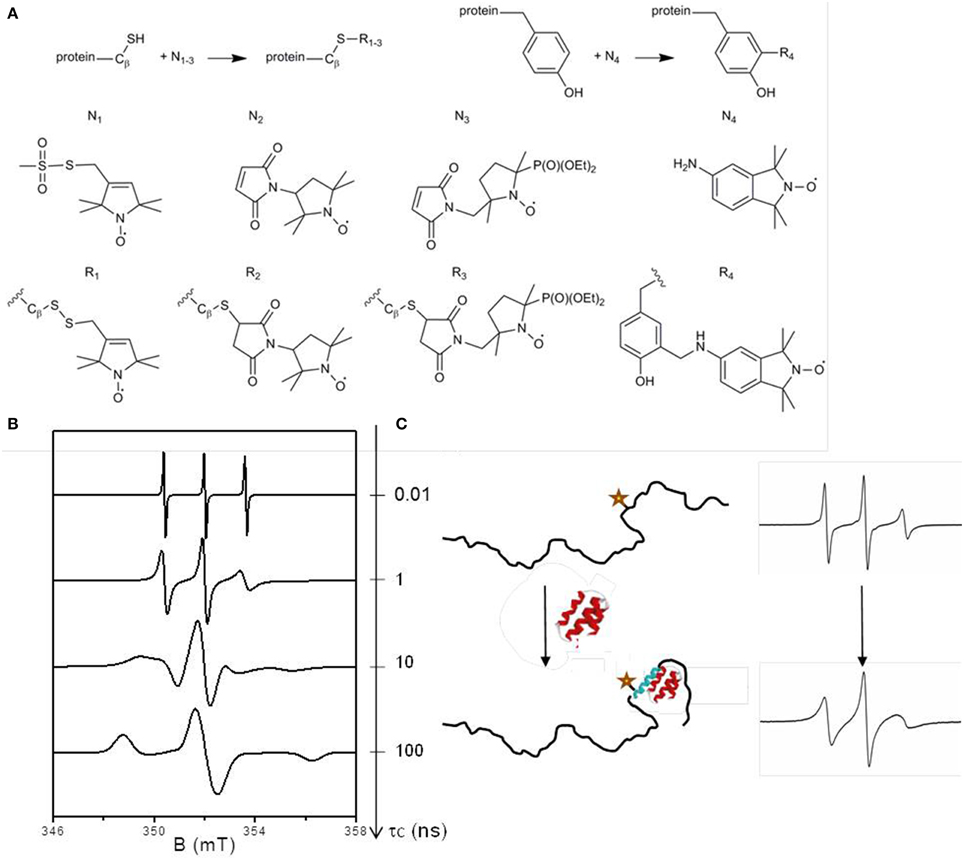
Structural Dynamics of Protein Interactions Using Site-Directed Spin Labeling of Cysteines to Measure Distances and Rotational Dynamics with EPR Spectroscopy

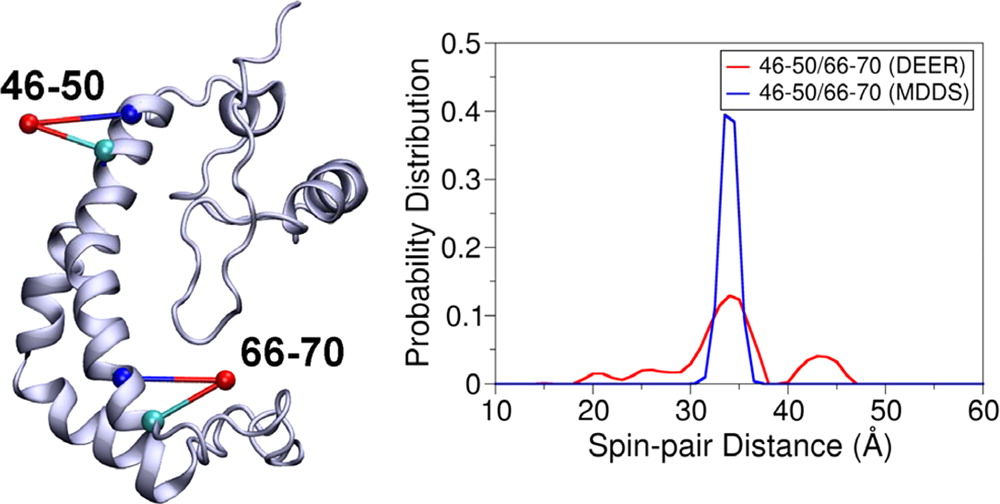
The main simulated distances (black) of PcrA expressed in Å between the

Interspin distance measurements. (A) Simulated powder spectra (obtained

Simulating the Distance Distribution between Spin-Labels Attached to Proteins

Site-directed spin labeling-electron paramagnetic resonance spectroscopy in biocatalysis: Enzyme orientation and dynamics in nanoscale confinement - ScienceDirect
chiLife: An open-source Python package for in silico spin labeling and integrative protein modeling
für dich empfohlen
 Fallbeispiel - Kohlensäure-Entfernung - EUWA14 Jul 2023
Fallbeispiel - Kohlensäure-Entfernung - EUWA14 Jul 2023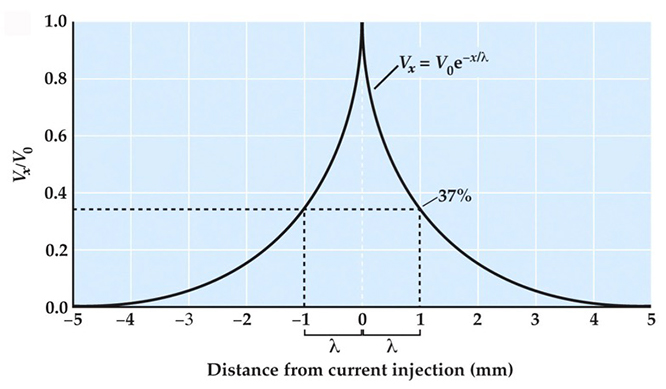 Neuroscience 6e Web Topic 2.1 - Passive Membrane Properties - Neuroscience, International Sixth Edition Student Resources - Learning Link14 Jul 2023
Neuroscience 6e Web Topic 2.1 - Passive Membrane Properties - Neuroscience, International Sixth Edition Student Resources - Learning Link14 Jul 2023- Impact of Operating Conditions on Measured and Predicted Concentration Polarization in Membrane Distillation14 Jul 2023
 Cell Membrane and Cell Transport - Worksheet14 Jul 2023
Cell Membrane and Cell Transport - Worksheet14 Jul 2023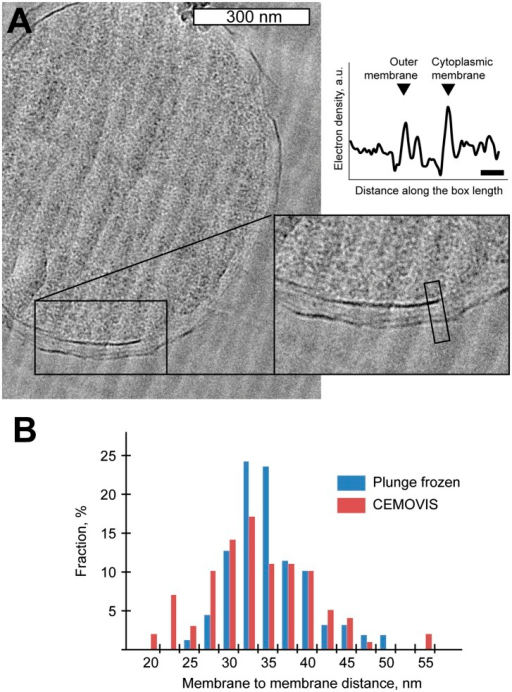 Comparison of membrane-to-membrane distance using CEMOV14 Jul 2023
Comparison of membrane-to-membrane distance using CEMOV14 Jul 2023 Cell Membrane and Transport - Google Slides and Distance Learning Ready14 Jul 2023
Cell Membrane and Transport - Google Slides and Distance Learning Ready14 Jul 2023 Universal 10 zoll Wasser Filter Patrone, UF Membran Für Wasser14 Jul 2023
Universal 10 zoll Wasser Filter Patrone, UF Membran Für Wasser14 Jul 2023 Neuroscience 6e Web Topic 2.1 - Passive Membrane Properties14 Jul 2023
Neuroscience 6e Web Topic 2.1 - Passive Membrane Properties14 Jul 2023 Membrane Protein Folding, Stability, and Protein-Membrane14 Jul 2023
Membrane Protein Folding, Stability, and Protein-Membrane14 Jul 2023 Nanostructure of the desmosomal plaque14 Jul 2023
Nanostructure of the desmosomal plaque14 Jul 2023
Sie können auch mögen
 ReiseAPOTHEKE rot - natur - Bestickend!14 Jul 2023
ReiseAPOTHEKE rot - natur - Bestickend!14 Jul 2023 Swing Deadpool Car Interior Decoration Rearview Mirror Pendant Car14 Jul 2023
Swing Deadpool Car Interior Decoration Rearview Mirror Pendant Car14 Jul 2023 Chrysler 300C CRD SRT Design (2008) driven review14 Jul 2023
Chrysler 300C CRD SRT Design (2008) driven review14 Jul 2023- Rammstein Belgium - RAMMSTEIN UNISEX PERFUME ”KOKAIN RED INTENSE” 100ML - RELOADED14 Jul 2023
 Tesla Badge Logo Emblem Sticker Covers for Tesla Model Y & Model 314 Jul 2023
Tesla Badge Logo Emblem Sticker Covers for Tesla Model Y & Model 314 Jul 2023 Maiqiken AntiDrop Autositz Lückenfüller 2 Stück 48cm Sitzlückenfüller Sitz Stopper für Golf Passat Tiguan : : Auto & Motorrad14 Jul 2023
Maiqiken AntiDrop Autositz Lückenfüller 2 Stück 48cm Sitzlückenfüller Sitz Stopper für Golf Passat Tiguan : : Auto & Motorrad14 Jul 2023 Motorrad Fighter 125 SR-S schwarz-rot Euro 5 125ccm Schaltmoped Leichtkraftrad14 Jul 2023
Motorrad Fighter 125 SR-S schwarz-rot Euro 5 125ccm Schaltmoped Leichtkraftrad14 Jul 2023 HIGH PRESSURE AIR TYRE INFLATOR FOR TRUCKS14 Jul 2023
HIGH PRESSURE AIR TYRE INFLATOR FOR TRUCKS14 Jul 2023 45° Alu-Bogen AD 25mm*** Alubogen Alurohr Aluminium Bogen14 Jul 2023
45° Alu-Bogen AD 25mm*** Alubogen Alurohr Aluminium Bogen14 Jul 2023 flexzon 12 V Auto Bus Truck Boot 5 Dixie Musik Auto Air Horn Dukes14 Jul 2023
flexzon 12 V Auto Bus Truck Boot 5 Dixie Musik Auto Air Horn Dukes14 Jul 2023

