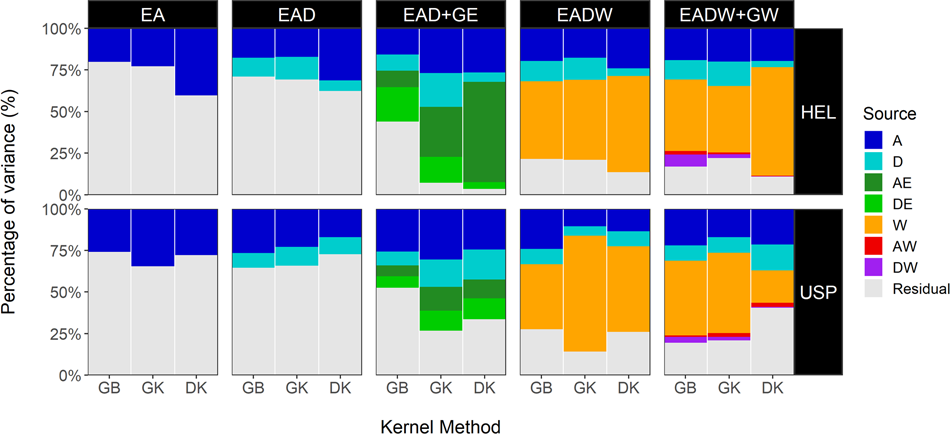
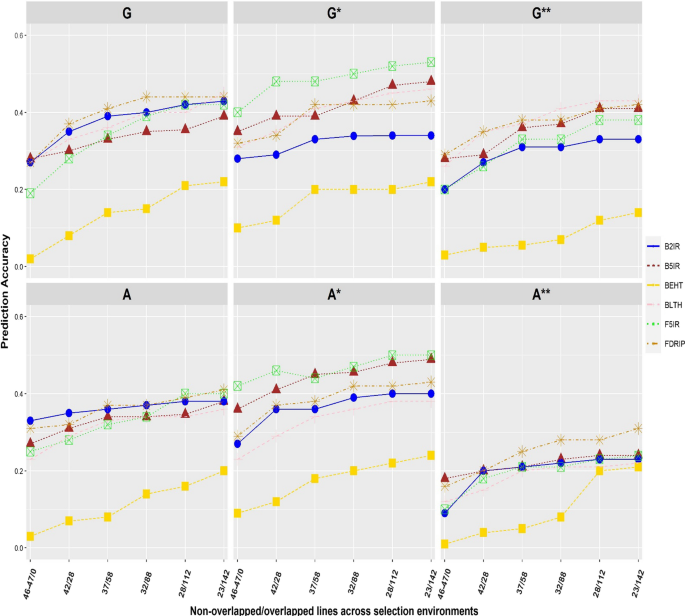
Nonlinear kernels, dominance, and envirotyping data increase the accuracy of genome-based prediction in multi-environment trials
Von einem Mystery-Man-Autor
Last updated 12 Juni 2024
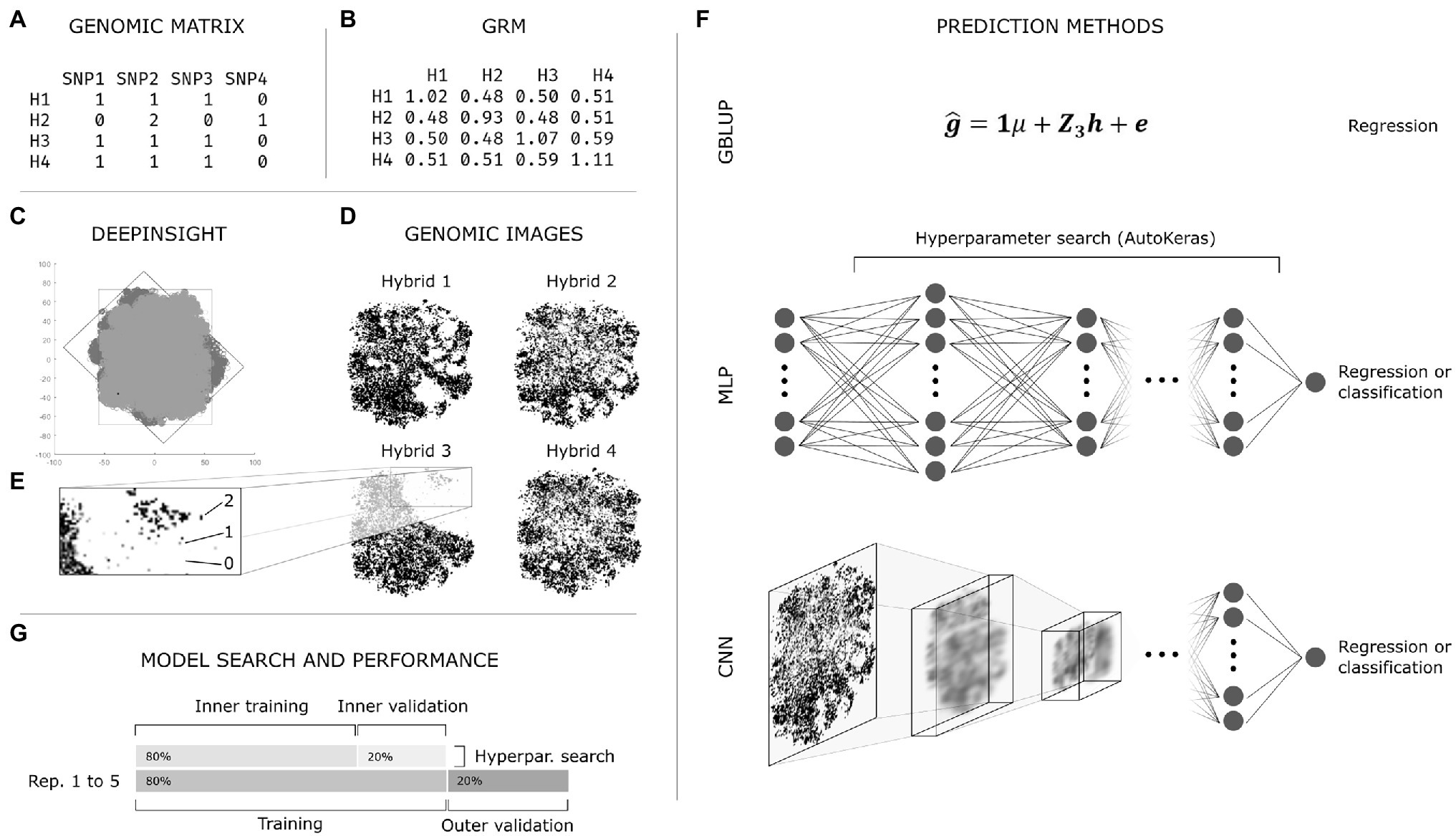
Frontiers Automated Machine Learning: A Case Study of Genomic “Image-Based” Prediction in Maize Hybrids
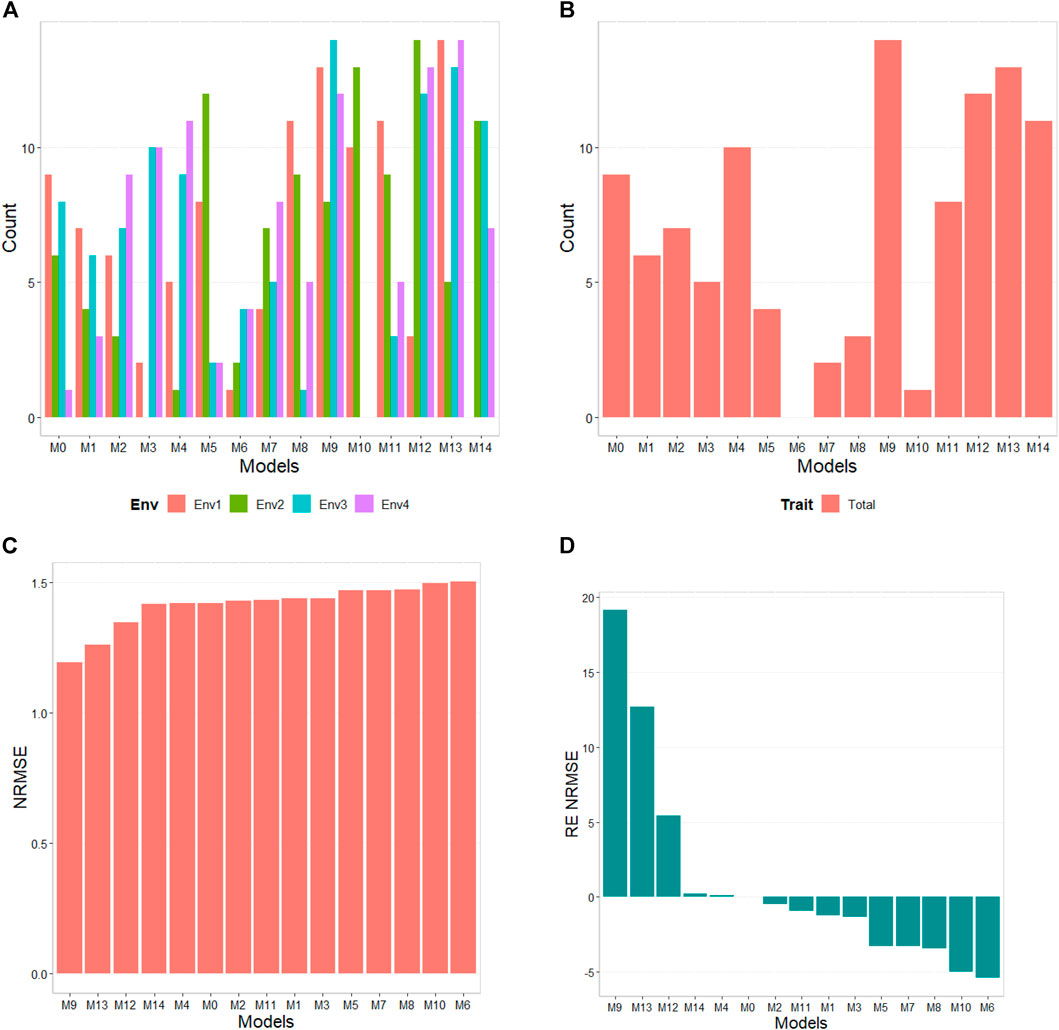
Frontiers Do feature selection methods for selecting environmental covariables enhance genomic prediction accuracy?

PDF) Nonlinear kernels, dominance, and envirotyping data increase the accuracy of genome-based prediction in multi-environment trials

Weighted Kernels Improve Multi-Environment Genomic Prediction
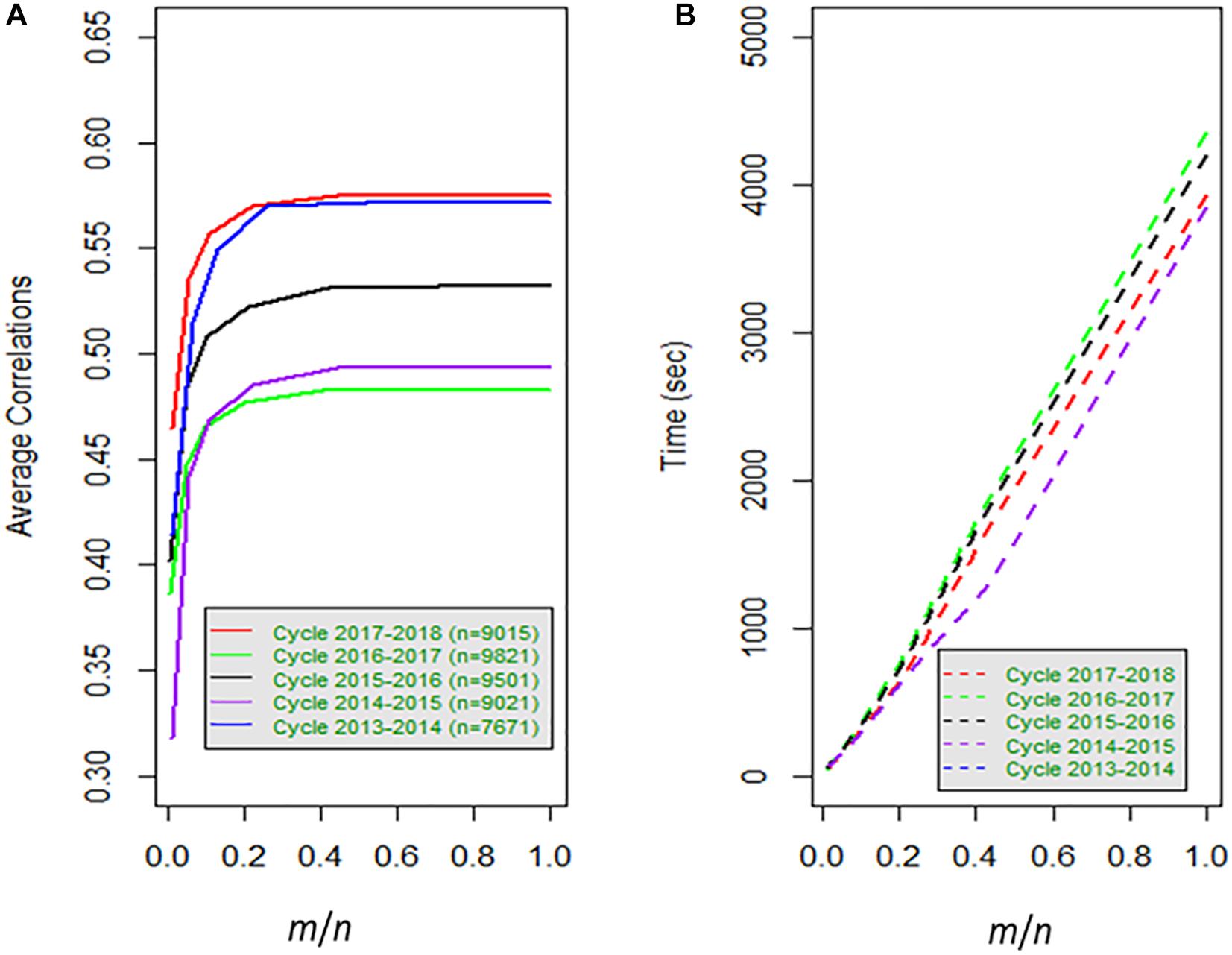
Frontiers Approximate Genome-Based Kernel Models for Large Data Sets Including Main Effects and Interactions
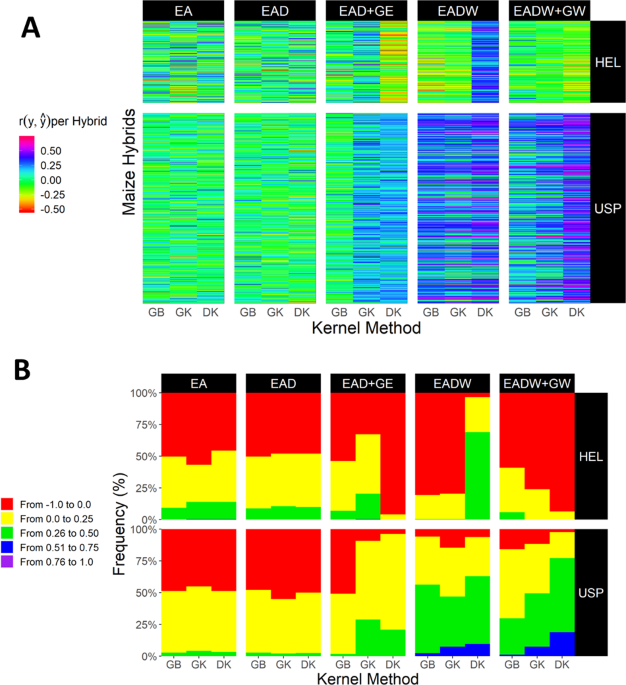
Nonlinear kernels, dominance, and envirotyping data increase the accuracy of genome-based prediction in multi-environment trials

Nonlinear kernels, dominance, and envirotyping data increase the accuracy of genome-based prediction in multi-environment trials
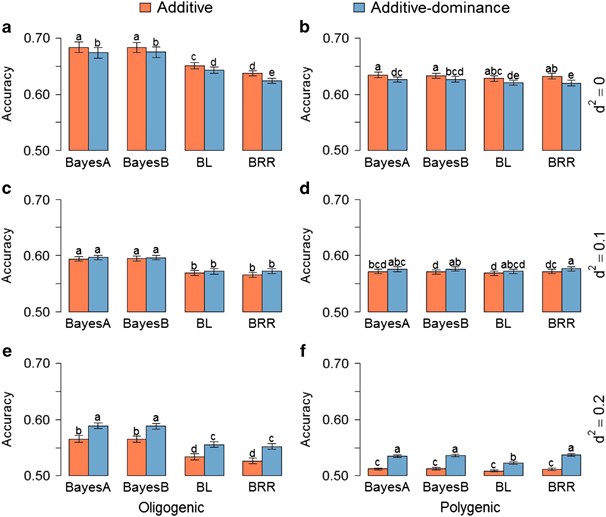
The contribution of dominance to phenotype prediction in a pine breeding and simulated population
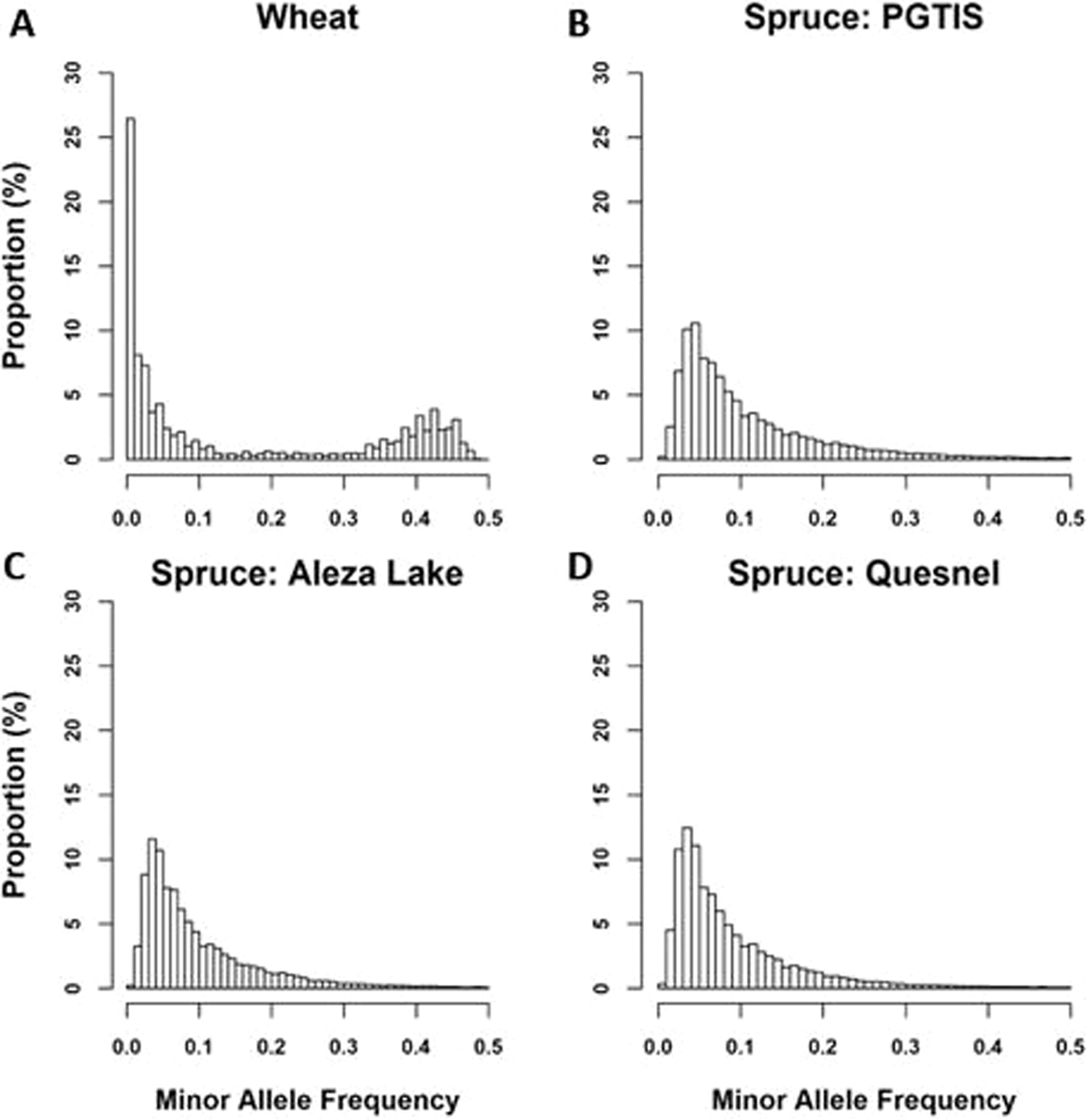
Weighted kernels improve multi-environment genomic prediction
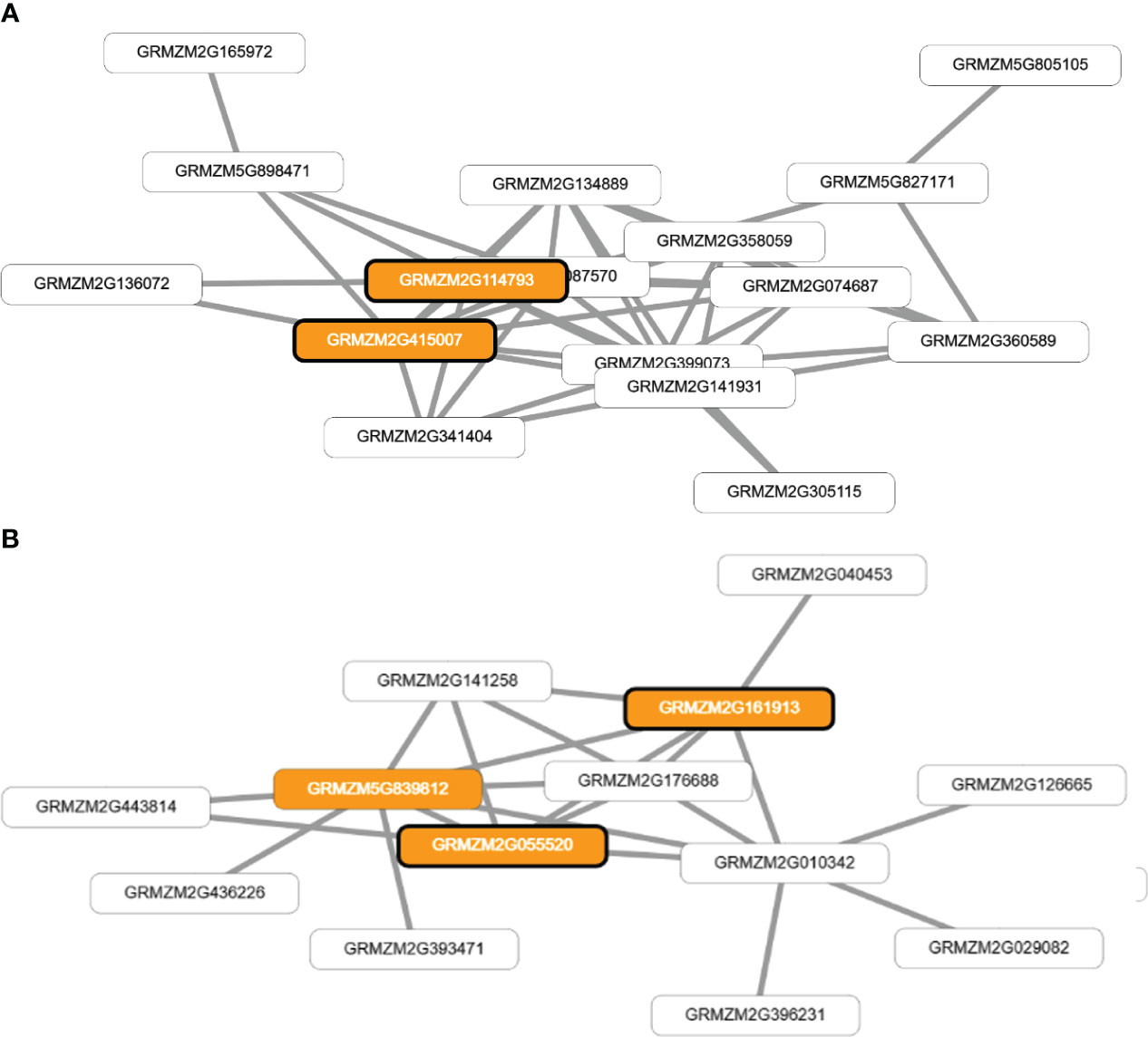
Frontiers Multi-trait and multi-environment genomic prediction for flowering traits in maize: a deep learning approach
Sparse testing using genomic prediction improves selection for breeding targets in elite spring wheat
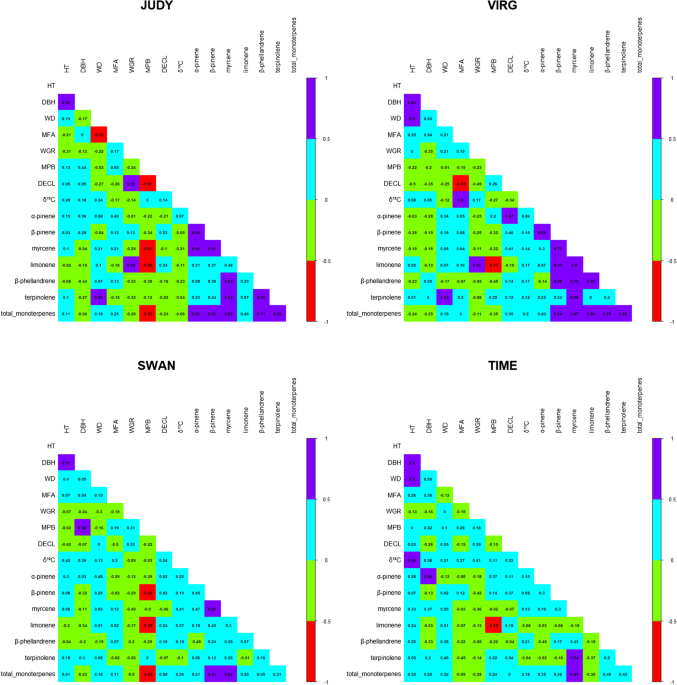
Multiple-trait analyses improved the accuracy of genomic prediction and the power of genome-wide association of productivity and climate change-adaptive traits in lodgepole pine, BMC Genomics
für dich empfohlen
 GK R 1/160/40G-FSTF14 Jul 2023
GK R 1/160/40G-FSTF14 Jul 2023 Genie Wireless Keyless Entry Keypad (GK-R) – The Genie Company14 Jul 2023
Genie Wireless Keyless Entry Keypad (GK-R) – The Genie Company14 Jul 2023 Most see him rollin', some be hatin': The Rota GKR14 Jul 2023
Most see him rollin', some be hatin': The Rota GKR14 Jul 2023 Führungswagen THK THK - ref. HSR-15-R-SS (CK-GK) - RUBIX Deutschland14 Jul 2023
Führungswagen THK THK - ref. HSR-15-R-SS (CK-GK) - RUBIX Deutschland14 Jul 2023 BOS Umfassungszarge GK125 Bud3 1-teilig BRM: 625x2125mm L/R für GK-Wand14 Jul 2023
BOS Umfassungszarge GK125 Bud3 1-teilig BRM: 625x2125mm L/R für GK-Wand14 Jul 2023 Zugkupplung Albe Berndes EM 80-R Ausf. GK , Gesamtgewicht 800 kg , Zugmaul . Kugelkupplung , Zugrohr rund 70 mm14 Jul 2023
Zugkupplung Albe Berndes EM 80-R Ausf. GK , Gesamtgewicht 800 kg , Zugmaul . Kugelkupplung , Zugrohr rund 70 mm14 Jul 2023 GKR Group Manufacturer of Plastic Injection Moulds14 Jul 2023
GKR Group Manufacturer of Plastic Injection Moulds14 Jul 2023 Top 600 General Knowledge Questions and Answers PDF Download: Enhance Your GK Knowledge with GKelite14 Jul 2023
Top 600 General Knowledge Questions and Answers PDF Download: Enhance Your GK Knowledge with GKelite14 Jul 2023 ASONPAO GK-R/GWKPD-BL Auto-Seek 315/390MHZ Keyless Entry Wireless Keypad 37224R for IntelliCode Genie Garage Door Opener(1Pack)14 Jul 2023
ASONPAO GK-R/GWKPD-BL Auto-Seek 315/390MHZ Keyless Entry Wireless Keypad 37224R for IntelliCode Genie Garage Door Opener(1Pack)14 Jul 2023 CM6321-R 12V DC 30A STARTER RELAY FOR GO KART GK 200D, F, H, E & MORE *OEM*14 Jul 2023
CM6321-R 12V DC 30A STARTER RELAY FOR GO KART GK 200D, F, H, E & MORE *OEM*14 Jul 2023
Sie können auch mögen
 3Pcs Auto Tür Fensterheber Schalter Taste Abdeckung Gehäuse Innen Dekoration Zubehör Für Benz Smart Fortwo Forfour 451 453 - AliExpress14 Jul 2023
3Pcs Auto Tür Fensterheber Schalter Taste Abdeckung Gehäuse Innen Dekoration Zubehör Für Benz Smart Fortwo Forfour 451 453 - AliExpress14 Jul 2023 orig. ANDRIS® Marken-Piktogramm Sicherheits-Aushang Schild „Verhalten bei Unfällen“, Erste Hilfe, Notfall, Hinweis, Betriebsaushang, ISO-Kunststoffplatte 200x200 mm14 Jul 2023
orig. ANDRIS® Marken-Piktogramm Sicherheits-Aushang Schild „Verhalten bei Unfällen“, Erste Hilfe, Notfall, Hinweis, Betriebsaushang, ISO-Kunststoffplatte 200x200 mm14 Jul 2023 Shop PSM Style Carbon Fiber Trunk Lip Spoiler Online14 Jul 2023
Shop PSM Style Carbon Fiber Trunk Lip Spoiler Online14 Jul 2023 BGS Ventilfederspanner Ventilfeder Spannapparat Ventil Federspanner 35 - 200 mm14 Jul 2023
BGS Ventilfederspanner Ventilfeder Spannapparat Ventil Federspanner 35 - 200 mm14 Jul 2023- LEICKE Blocker-Karten-Set RFID Karte(für Universal meisten Kreditkarten (VISA, Mastercard), EC-Karten, Scheckkarten, Führerschein- und Mitgliedskarten, Türschloss-Karten, Personalausweise, Reisepässe, Chipkarten, Zugangskarte, Bankkarten)14 Jul 2023
 Magnetplatten - Buchbindermeister2414 Jul 2023
Magnetplatten - Buchbindermeister2414 Jul 2023 PopMount 2 Auto und Tisch Schwarz Halterung14 Jul 2023
PopMount 2 Auto und Tisch Schwarz Halterung14 Jul 2023 Caliber Audio Technology RMD120DAB-BT-B Autoradio Bluetooth MP314 Jul 2023
Caliber Audio Technology RMD120DAB-BT-B Autoradio Bluetooth MP314 Jul 2023 Hazet Universal Kugelgelenk-Abzieher 1790-714 Jul 2023
Hazet Universal Kugelgelenk-Abzieher 1790-714 Jul 2023 Silikon Hülle kompatibel mit Apple iPhone 13 Mini Case transparent Handyhülle Cars Disney Pixar Lightning McQueen 95: : Elektronik & Foto14 Jul 2023
Silikon Hülle kompatibel mit Apple iPhone 13 Mini Case transparent Handyhülle Cars Disney Pixar Lightning McQueen 95: : Elektronik & Foto14 Jul 2023
